ANOVA
What, and why?
Analysis of variance (ANOVA) is a statistical technique that lets us compare means of different groups of data to determine whether they differ significantly.
At its most basic, a one-way ANOVA (i.e., with only one factor) with only two groups is the same as a t-test. ANOVA scales up this analysis, allowing for more than two groups within a factor, and for multiple factors.
Under the hood, ANOVA is really a linear model where the explanatory variable is categorical rather than continuous. This is made clear by the process of running ANOVA in R. The basic approach is to use the function aov, which fits an ANOVA model by a call to lm. Alternatively, you can use lm directly. The output varies somewhat when the model is passed to the summary or print functions, but the statistical analysis is the same.
One-way ANOVA
We’ll start with a one-way ANOVA, using the Inverts data from the previous few lessons. We have data from multiple catchment types (Type): agricultural, forest, and urban. Our question is whether invertebrate species richness varies with catchment type.
To run an ANOVA, we need our explanatory variable to be a factor (i.e., rather than a character string). If you don’t have Inverts in your environment, read it in, and make sure that stringsAsFactors is set to TRUE. If you already have it in your environment, but Type and Country are characters, you can use mutate and as.factor to convert them to factors.
# Read in Inverts data (first introduced in functions lesson)
Inverts <- read.csv(file="Data/Inverts.csv", stringsAsFactors=TRUE, header=TRUE)
# Or, if Inverts is already in your environment with Type and Country as characters,
# convert them to factors
Inverts <- Inverts %>% mutate(Type = as.factor(Type), Country = as.factor(Country))Look at your data!
Once again, we will start by looking at our data! Let’s see if there is any evidence for richness varying with catchment type. We can do this with the plot function, which acts similarly to boxplot when we are plotting continuous data against factors.
plot(Richness ~ Type, data=Inverts)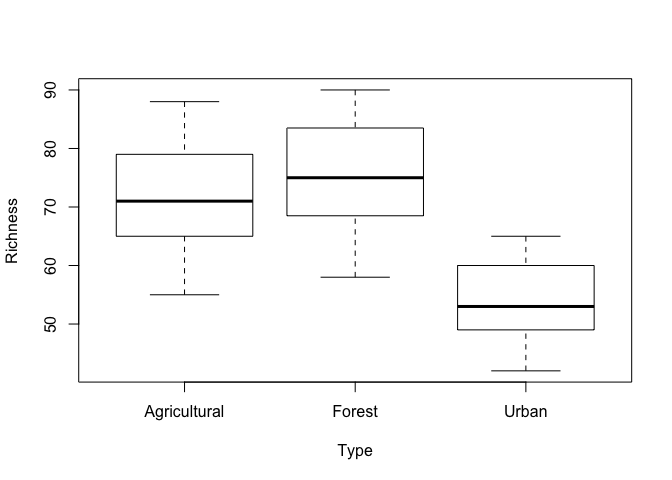
It looks like invertebrate species richness is fairly comparable between streams in agricultural and forested areas, but lower in urban areas. However, there is enough variation that we don’t know if this will be a statistically significant difference. So let’s run an ANOVA and find out.
Fit the model
To fit an ANOVA to the data, we can use either lm or aov, which will effectively do the same thing. The main difference is how the output is presented in a call to summary. Let’s use aov, which will put the output of summary in a form that makes sense for ANOVA results.
The help file for aov (?aov) notes that the default contrasts in R are not orthogonal, and aov will work better with orthogonal contrasts, so let’s start by setting them. Contrasts have to do with how the levels of each factor are compared to each other in a linear model. Information on the various contrasts available in R can be found in the help file for specific contrasts (e.g., ?contr.helmert), and more detail on how each contrast setting works can be found in this post on unordered contrats and a UCLA Stat Consulting Group page.
The following code both sets the contrasts to be orthogonal (contr.helmert for unordered factors and contr.poly for ordered factors) and assigns the original options to op so that they can be re-set using options(op). (You can also set the contrasts within the call to lm or aov later on, using the argument contrasts, but instead we will do it at the beginning for convenience - it will now apply to all downstream models.)
# Set default values to op to be able to re-set later;
# set current values to contr.helmert for unordered factors and contr.poly for ordered factors.
op <- options(contrasts = c(unordered="contr.helmert", ordered="contr.poly"))
# Check contrasts in current settings, if you'd like
options()$contrasts## unordered ordered
## "contr.helmert" "contr.poly"Now we can fit the model. We’ll store it as a named object so that we can work with it more easily later.
You can name this object however you want, but it’s good to be consistent with your naming scheme throughout your code, so that you can easily identify what kind of object you are working with. There are multiple naming conventions used in R and R packages, as well as multiple style guides (e.g., see here). In the case below, the . in the model name is only for convenient reading. (There is some debate on when and how this . should be used. This is in part because the . has a specific meaning in other programming languages, whereas it is simply another character in an R variable name.)
InvertsType_mod <- aov(Richness ~ Type, data=Inverts)Now let’s take a look at the model object with summary.
summary(InvertsType_mod)## Df Sum Sq Mean Sq F value Pr(>F)
## Type 2 2094 1046.8 11.23 0.000483 ***
## Residuals 21 1958 93.3
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1This output gives us the results of an F-test with the null hypothesis that the groups are from the same statistical population, which would mean that their means do not vary. We can see that p<0.001, indicating that the groups are likely from different populations. Thus, species richness significantly varies with catchment type.
NOTE: The ANOVA table given by summary with an aov model gives results using Type I SS (Sum of Squares). The function anova gives the same output as summary, and similarly uses Type I SS. This does not matter for a one-way ANOVA, but it absolutely does with multiple factors! We will cover this further in the Two-way ANOVA section. For now, the point to remember is that you should not accidentally report your results from summary on an ANOVA model unless you are doing a one-way ANOVA, or if your experimental question is such that Type I SS makes sense (which is not common).
Check model assumptions
Of course, we can’t use this model unless we know that we have satisfied the model’s assumptions! As with linear regression, we need to check the homogeneity and normality of the residuals. We will make the plots as before, starting with a histogram and a QQ plot of the residuals, and then comparing the residuals to the explanatory variable, Type.
par(mfrow=c(2,2)) # Set plot layout
hist(resid(InvertsType_mod), las=1) # Look at distribution of residuals
qqnorm(resid(InvertsType_mod), las=1) # Compare distribution of residuals to a normal distribution
qqline(resid(InvertsType_mod))
plot(fitted(InvertsType_mod), resid(InvertsType_mod), las=1) # Do residuals vary by fitted values?
plot(Inverts$Type, resid(InvertsType_mod), las=1) # Do the residuals vary by group?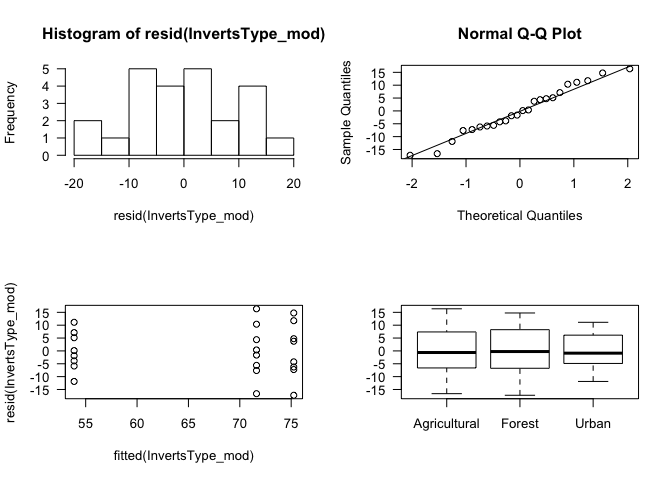
par(mfrow=c(1,1)) # Re-set plot layoutThese plots give us good news. The distribution of residuals doesn’t look extraordinarily normal in the histogram, but this could be an effect of small sample size, and the QQ plot suggests a mostly normal distribution with a few potentially outlying points. The bottom plots suggests that the residuals are comparable among groups (homogeneity). On the plot of the residuals versus the fitted values, the values on the x-axis might seem obscure at first - these are the means for the three groups (Agricultural, Forest, and Urban).
We can also make similar plots by making a function call to plot using the model as input. In this case, the function will automatically point out potential outliers.
par(mfrow=c(2,2)) # Set plot layout for 4 plots
plot(InvertsType_mod, las=1) # Plot residuals for our ANOVA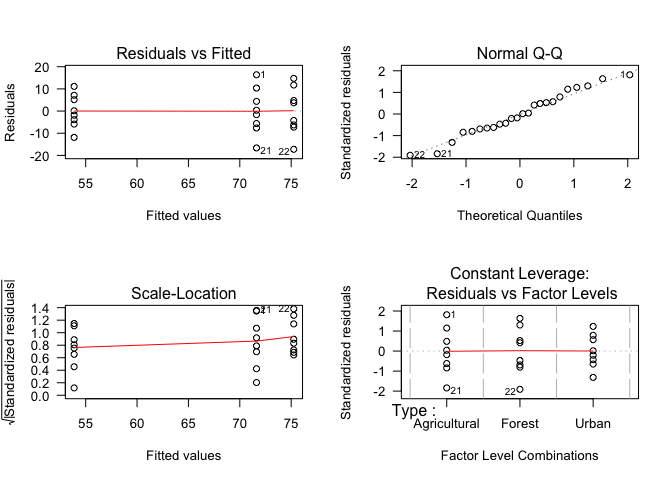
par(mfrow=c(1,1)) # Re-set plot layoutNow we can see that there are three points that are potential outliers. If these were real data, it could be useful to examine these specific points further to determine whether there is any reason not to include them in the analysis.
These plots indicate that out model is appropriate for these data, and we can report the results. Note, however, that the opposite result - e.g., discovering that the residuals are not normally distributed - can still be immensely useful, as visualizing the residuals (and their distribution) can help you determine what would be an appropriate approach. For example, if you saw that your residuals were right-skewed, this would suggest that log-transformation of the dependent variable (before fitting the ANOVA) would result in normally distributed residuals.
Post-hoc testing
We’ve interpreted the results to mean that species richness varies with catchment type. But, which catchment types differ from each other? To assess this, we can use a Tukey test with the function TukeyHSD (for Tukey’s “Honestly Significant Difference” method).
TukeyHSD(InvertsType_mod)## Tukey multiple comparisons of means
## 95% family-wise confidence level
##
## Fit: aov(formula = Richness ~ Type, data = Inverts)
##
## $Type
## diff lwr upr p adj
## Forest-Agricultural 3.625 -8.545074 15.795074 0.7364753
## Urban-Agricultural -17.750 -29.920074 -5.579926 0.0038490
## Urban-Forest -21.375 -33.545074 -9.204926 0.0006580The top part of the output tells us that we’re using 95% confidence intervals. If for some reason we wanted to change this, we could use the argument conf.level, which defaults to 0.95.
The table below $Type gives us the p-values for comparisons between groups. From this, we can see that at a significance level of 0.05 (\(\alpha\) = 0.05), species richness in forested and agricultural areas are comparable, whereas that of urban areas differs significantly from the two other groups.
Challenge
Read in the
treesdata set that we used in the lesson on reshaping data frames with dplyr. Across all sites, does tree abundance (Count) vary by species? If so, which species differ? Whether or not the model is significant, check whether the model assumptions are met.The file
allbirds.csvcontains count data on 24 bird species that were observed at one site during 4 different months. Read in this data, and use an ANOVA to determine whether species counts varied by month. If so, run a post-hoc test to determine which months vary. Use a boxplot to assess data visually, and be sure to check model assumptions!
Two-way ANOVA
We also have data on the country for each stream sample, and we might want to know whether invertebrate species richness varies between the US and Canada as well as catchment type. In addition, we might want to know whether the relationship between species richness and catchment type varies based on country. (This could be a biogeographical effect - or maybe invertebrates have sociopolitical preferences?!) To look at both of these factors, Type and Country, we can use a two-way ANOVA.
Look at your data (again)!
We will of course start by looking at the data. We can look at species richness by country, to complement the above plot of species richness by catchment type, or we can look at species richness by all country-type groups, using boxplot.
p <- par(mar=c(10,4,2,2)) # Save current parameters & re-set margins to allow for long x-axis labels
boxplot(Richness ~ Type + Country, data=Inverts, las=2)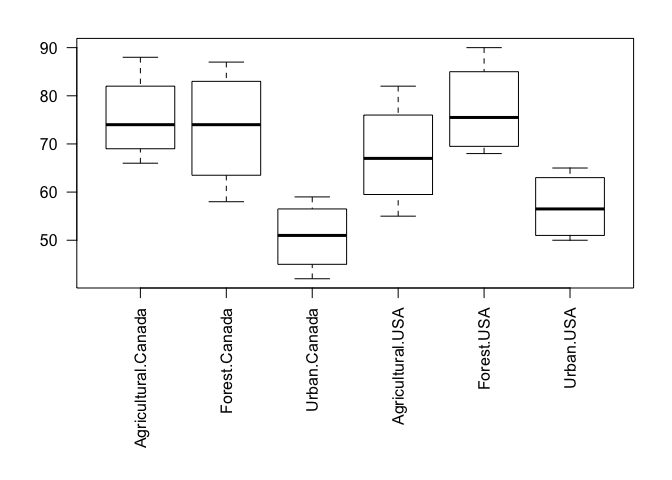
par(p) # Reset marginsWe can still see the differences between catchment type, but it’s difficult to tell whether there are differences by country, especially between urban catchments. So, let’s add Country to our model to see if it has an effect.
Fit the model
Implementing a two-way ANOVA is similar to implementing ANOVA, in that we will use the model function (aov). It is also similar to multiple regression, in terms of the way that we specify multiple factors or explanatory variables. Here, we will use * to include both main effects and their interaction.
Inverts2way_mod <- aov(Richness ~ Type * Country, data=Inverts)
summary(Inverts2way_mod) # F-test with Type I Sum of Squares (probably not appropriate!)## Df Sum Sq Mean Sq F value Pr(>F)
## Type 2 2093.6 1046.8 10.904 0.00079 ***
## Country 1 4.2 4.2 0.043 0.83731
## Type:Country 2 226.1 113.0 1.178 0.33068
## Residuals 18 1728.0 96.0
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Here is where it is important to remember that summary gives you results using Type I Sum of Squares! So, do not stop here to determine the significance of the independent variables.
Sidenote: Type I vs Type III Sums of Squares
Differences between different Types of Sums of Squares have to do with how the significance of each factor (independent variable or interaction) is considered relative to the others in the model. The importance of a given factor to predicting the dependent variable can be assessed by comparing the sum of squares when of the model including this factor to the sum of squares of the model without it.
With Type I SS, the first factor on the right side of the formula is added to the model first, and the sum of squares is calculated. Then the next variable (or interaction) is added, and the change in sum of squares is calculated. And so on, and so forth. For a two-way ANOVA with factors A and B and the interaction A:B, this means the main effect of factor A is calculated first, then the main effect of factor B given the main effect of factor A, and last the interaction of A and B after the main effects. This is why type I SS is also called “sequential sum of squares.” This means that the results depend on the order in which the main effects are given in the model.
With Type III SS, the significance of each variable is assessed by comparing a model with all effects and interactions except the variable of interest to the same model with the variable of interest. So the question is, given everything else in the model, do we increase explanatory power by including this variable? This process is repeated for each variable and interaction separately.
Your choice of model depends on your experimental question. When you’re interested in the effects of both factors in ecology, you’re generally interested in the effect of a factor after controlling for the others, in which case type III is what you should choose.
For more information, this blog post has a nicely formatted version of Falk Scholer’s explanation, and the top response to this CrossValidated post on StackExchange had a nice explanation of how sums of squares are partitioned between effects in the different tests.
…back to our model
If we want the results of an F-test using type III SS, we have several options - a good reminder that there are often multiple ways of doing the same thing in R! One option is to use the drop1 function with an F test, which compares refitted models with different terms dropped. Another option is to use the Anova function in the package car, and specify Type = III. Both approaches will give you the same results for linear models, with only slightly different output due to their methods: drop1 will give you AIC scores for the different models compared, and Anova will show you the significance of the intercept. If you don’t need either of these, choose the function you like better - e.g., based on which syntax feels more intuitive to you, or whether you have installed the car package (but, you can run install.packages("car") if not)1. We will show that both give the same statistics.
# With drop1, the scope argument designates the terms to be dropped; the . means all of them.
drop1(Inverts2way_mod, scope = ~., test="F") ## Single term deletions
##
## Model:
## Richness ~ Type * Country
## Df Sum of Sq RSS AIC F value Pr(>F)
## <none> 1728.0 114.64
## Type 2 2093.58 3821.6 129.69 10.9041 0.0007901 ***
## Country 1 4.17 1732.2 112.70 0.0434 0.8373079
## Type:Country 2 226.08 1954.1 113.59 1.1775 0.3306785
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1library("car")##
## Attaching package: 'car'## The following object is masked from 'package:dplyr':
##
## recodeAnova(Inverts2way_mod, type="III") # Specify Type III SS## Anova Table (Type III tests)
##
## Response: Richness
## Sum Sq Df F value Pr(>F)
## (Intercept) 107468 1 1119.4601 < 2.2e-16 ***
## Type 2094 2 10.9041 0.0007901 ***
## Country 4 1 0.0434 0.8373079
## Type:Country 226 2 1.1775 0.3306785
## Residuals 1728 18
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1With either function, we get the p-value for the F test with the null hypothesis that the different groups are from the same statistical population and thus have the same means. Here, p<0.05 for catchment Type! So this indicates that there are significant differences in species richness in streams from different catchment types.
As a sidenote, here is where we could consider dropping the interaction term, since it is not significant. However, we will move on for now.
Check assumptions
To use this model, we need to check that its assumptions have been met. We will look at the model residuals as before.
par(mfrow=c(2,2)) # Set plot layout
plot(Inverts2way_mod, las=1)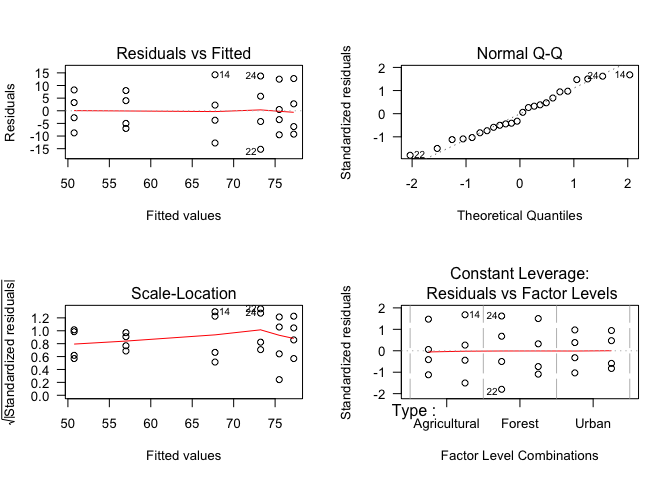
par(mfrow=c(1,1)) # Re-set plot layoutAnd as before, our residuals look fairly normal and homogeneous, so we can report the model output.
Post-hoc testing
We may still wonder about the differences between levels of the significant factor, Type. With a 2-way ANOVA, we can apply a Tukey test using the function lsmeans in the package lsmeans, along with the function cld, which controls the output that is displayed. The first argument to cld is an object created by lsmeans. lsmeans requires us to specify the model as well as the variable to test (e.g., Type), and give us confidence intervals as output, which we could use to determine which groups differ. We can also use cld to figure this out for us. For cld, we can to specify the p-value as alpha, the post-hoc test to use as adjust, and whether to use letters instead of the default to numbers for comparing groups.
If you have not already done so, be sure to install lsmeans and associated dependencies, i.e. with install.packages("lsmeans", dependencies = TRUE).
library("lsmeans")## Loading required package: estimability# Which Types vary from the others, using a Tukey test?
cld(lsmeans(Inverts2way_mod, specs="Type"), alpha=0.05, adjust="tukey", Letters=letters) ## NOTE: Results may be misleading due to involvement in interactions## Type lsmean SE df lower.CL upper.CL .group
## Urban 53.875 3.464102 18 44.76064 62.98936 a
## Agricultural 71.625 3.464102 18 62.51064 80.73936 b
## Forest 75.250 3.464102 18 66.13564 84.36436 b
##
## Results are averaged over the levels of: Country
## Confidence level used: 0.95
## Conf-level adjustment: sidak method for 3 estimates
## P value adjustment: tukey method for comparing a family of 3 estimates
## significance level used: alpha = 0.05Take a look at the output under .group. Here we can see that the Urban streams have significantly different invertebrate species richness than the other catchment types. And, this approach gives us the added convenience of letters that can be used to denote significance on a figure!
If the interaction were significant and we wanted to do pairwise comparisons between groups, we could use the lsmeans function with specs = pairwise ~ Type * Country. You can try this out if you’re curious about the output.
So it looks like we can conclude that (in the world represented by this fake data) species richness varies among catchment types, with lower richness in urban areas - but we can only conclude this if the model assumptions are met!