Control structures 2: for loops
Why, and what?
Control structures allow us to alter the flow of a program, and to make decisions about when or how many times an action occurs. These tools can help you automate processes that require decision-making or repeated actions.
Here we will introduce you to the second of two control structures covered in this course: for loops.
for loops
Another useful control structure that we can use in R is a for loop. For loops iterate over values in a vector, and execute code for each value. This can be extremely helpful if you want to perform an action multiple times! For example, you might want to read in multiple files, or do the same thing to multiple data tables. The flow of a for loop looks like this:
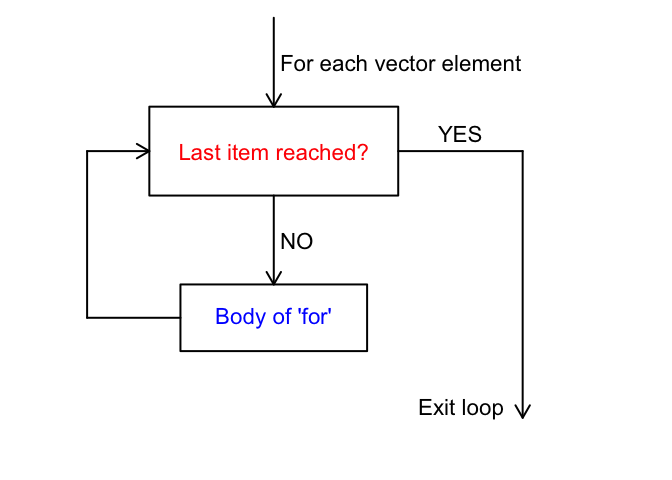
The code in R has this general structure:
for (n in c(your vector)) {
do this thing involving n
}In this loop, n will be set iteratively to each of the values in your vector, and the code in brackets will be executed for each value of n. To demonstrate how this works, let’s print every number in a vector.
for (n in c(1:5)) {
print(n)
}## [1] 1
## [1] 2
## [1] 3
## [1] 4
## [1] 5It’s not important what variable we put in the place of n. We can name this variable x, or trees, or fabulousvariable, or Constantinople.
for (Constantinople in c(1:5)) {
print(Constantinople)
}## [1] 1
## [1] 2
## [1] 3
## [1] 4
## [1] 5We can even create a vector of character strings, and have the for loop step through each element of this character vector.
for (colors in c("red", "blue", "green", "yellow", "orange", "white")) {
print(colors)
}## [1] "red"
## [1] "blue"
## [1] "green"
## [1] "yellow"
## [1] "orange"
## [1] "white"And, we can do whatever we want with the variable inside the for loop. For example, we can multiply the value of a numeric variable by 2, or we could put a character variable into a sentence.
# Multiply each element of a numeric vector by 2, and print this new value
for (Constantinople in c(1:5)){
print(Constantinople * 2)
}## [1] 2
## [1] 4
## [1] 6
## [1] 8
## [1] 10# Print a sentence with each element of a character vector
# (Use paste to put the character strings into one)
for (colors in c("red", "blue", "green", "yellow", "orange", "white")) {
print(paste("I am sure that",colors,"is the best color of all!"))
}## [1] "I am sure that red is the best color of all!"
## [1] "I am sure that blue is the best color of all!"
## [1] "I am sure that green is the best color of all!"
## [1] "I am sure that yellow is the best color of all!"
## [1] "I am sure that orange is the best color of all!"
## [1] "I am sure that white is the best color of all!"But why would I want to use a for loop, you ask?
For loops can be useful for applying the same action to multiple objects. For example, let’s say that you have many data frames with the variable “Species_Names”, and you want to rename that column to simply be “Species.” Let’s read in files like this, with bird count data from several different months.
# Read in bird count data
birds_jan <- read.csv(file="Data/birds_jan.csv")
birds_feb <- read.csv(file="Data/birds_feb.csv")
birds_mar <- read.csv(file="Data/birds_mar.csv")
birds_apr <- read.csv(file="Data/birds_apr.csv")
head(birds_jan)## SpeciesNames Count
## 1 Haemorhous purpureus 58
## 2 Sturnus vulgaris 48
## 3 Accipitridae sp. 39
## 4 Dryocopus pileatus 54
## 5 Mergus merganser 46
## 6 Phasianus colchicus 37You can rename a variable in a data frame using the function rename in dplyr. Note that within the function, the syntax is NewName = OldName.
# Rename variable from "SpeciesNames" to "Species"
birds_jan <- rename(birds_jan, Species = SpeciesNames)
head(birds_jan)## Species Count
## 1 Haemorhous purpureus 58
## 2 Sturnus vulgaris 48
## 3 Accipitridae sp. 39
## 4 Dryocopus pileatus 54
## 5 Mergus merganser 46
## 6 Phasianus colchicus 37# Reset original name
birds_jan <- rename(birds_jan, SpeciesNames = Species)Alternatively, we can do this with assign to be more concise. assign is exactly like <-, except that the variable name and the value are specified inside the function.
# Rename variable from "SpeciesNames" to "Species"
assign(x="birds_jan", value=rename(birds_jan, Species = SpeciesNames))
head(birds_jan)## Species Count
## 1 Haemorhous purpureus 58
## 2 Sturnus vulgaris 48
## 3 Accipitridae sp. 39
## 4 Dryocopus pileatus 54
## 5 Mergus merganser 46
## 6 Phasianus colchicus 37# Reset original name
birds_jan <- rename(birds_jan, SpeciesNames = Species)But what if we want to change the variable name in every single data frame? With four data frames, it might not be terrible to write out a line for each one, but with more data frames, this could quickly become an unwieldy time sink.
Enter for loops. With for loops, we can approach the problem differently. First, we will create a vector with the names of all the data frames. Then, we will use a for loop to step through every element of that vector - i.e., each data frame name - and change the variable name for each one.
# Create a vector with the names of all the data frames we want to work on
dfnames <- c("birds_jan", "birds_feb", "birds_mar", "birds_apr")
# Step through the above vector and rename the SpeciesNames variable to Species
for (dataframe in dfnames){
assign(x = dataframe, value = rename(get(dataframe), Species = SpeciesNames))
}Code trick sidenote: In the above code, we needed to use the function get to specify the data frame in rename. This is because what we have is a character vector that is the name of the data frame (e.g., "birds_jan"), but rename is looking for the data frame itself (e.g., birds_jan). get gets the value of the variable that you pass to it.
To explore this, try typing dfnames[1], and then try typing get(dfnames[1]).
The output of the first one is the first element of dfnames, which is the name of the data frame. The second one, however, gives us the value of the data frame that has that name. get searches for an object with the name specified by the x argument. This allows you to pass a vector or list of names of variables to other functions.
As another example, try running x <- 5, then try "x" versus get("x").
And now we can use another for loop to look at the results of our code!
for (dataframe in dfnames){
dataframe %>% print() # Print name of data frame to screen
get(dataframe) %>% head() %>% print() # Get data frame from its name, show head, print to screen
}
## Note that we could also have set this up with a numeric vector of 1:4,
## instead of the vector dfnames:
# for (n in 1:length(dfnames)){
# dfnames[n] %>% print()
# get(dfnames[n]) %>% heads() %>% print()
# }## [1] "birds_jan"
## Species Count
## 1 Haemorhous purpureus 58
## 2 Sturnus vulgaris 48
## 3 Accipitridae sp. 39
## 4 Dryocopus pileatus 54
## 5 Mergus merganser 46
## 6 Phasianus colchicus 37
## [1] "birds_feb"
## Species Count
## 1 Regulus satrapa 27
## 2 Zonotrichia albicollis 40
## 3 Eremophila alpestris 2
## 4 Larus delawarensis 18
## 5 Spinus tristis 12
## 6 Picoides villosus 47
## [1] "birds_mar"
## Species Count
## 1 Cathartes aura 16
## 2 Plectrophenax nivalis 9
## 3 Junco hyemalis hyemalis/carolinensis 58
## 4 Accipitridae sp. 12
## 5 Icterus bullockii/galbula 18
## 6 Poecile atricapillus 52
## [1] "birds_apr"
## Species Count
## 1 Regulus satrapa 25
## 2 Poecile hudsonicus 49
## 3 Spinus tristis 37
## 4 Columba livia (Feral Pigeon) 13
## 5 Plectrophenax nivalis 27
## 6 Zonotrichia albicollis 18Another example: Reading in multiple files
A slightly more complicated, but potentially very useful, example could be if you want to read in multiple data files automatically and assign them to different data frames. To do this, we can start with a data frame specifying the names of the files and the data frame names that we want to give to them.
ExperimentFiles <- data.frame(files = c("Data/Experiment_carbon.csv", "Data/Experiment_nutrients.csv")
,names = c("carbon", "nutrients")
,stringsAsFactors = FALSE)
ExperimentFiles## files names
## 1 ../Data/Experiment_carbon.csv carbon
## 2 ../Data/Experiment_nutrients.csv nutrientsNow we can step through the observations in ExperimentFiles, read in each file, and assign it to a data frame with a specified name.
for (n in 1:2) {
assign(x = ExperimentFiles$names[n], value = read.csv(file = ExperimentFiles$files[n]))
get(x = ExperimentFiles$names[n])
}There are several layers to this piece of code. The assign function assigns the value specified by value to a variable named x. When n is 1, the name of the variable will be first observation 1 in ExperimentFiles$names, and the value of the variable is the .csv file that we’re reading in, specified by observation 1 in ExperimentFiles$files. When n is 2, the name and value of the variable will be the second observations in our data frame.
The last line in the for loop uses the get function, as above. Try typing ExperimentFiles$names[n] without the get function.
The output is the nth character in that variable, i.e. "carbon" or "nutrients". What we want, though, is the value of the variable that has that name. To do that, we use the get function, which searches for an object with the name specified by the x argument. This allows you to pass a list of names of variables to other functions.
Challenge
Write a for loop that steps through a numeric vector and prints a number that is 4 times the number of the numeric vector.
The code within the brackets
{ }of a for loop does not necessarily have to refer to the values in the controlling vector! The controlling vector can be there just to determine the number of times to go through the loop. Write a for loop that loops through 10 times, and each time, adds 1 to a given variable (specified before the for loop) and then prints the value of that variable. (I.e, the variable should increase in value with each loop.)Write a for loop that goes through the rows of the
treesdata frame and prints the value ofCountif theSpeciesisAcer rubrum. In this case, the for loop should be used to indicate which row oftrees(or, which element of a variable withintrees), to examine.- Write a for loop that goes through the
nutrientsdata frame and prints the value of eachNitriteobservation plus 1. As above, the for loop effectively indicates which row to examine.- Next, create an empty vector called
nitrite_plus. (You can do this withc().) Then, modify your for loop so that it adds each “Nitrite plus 1” value to the end of this vector.
- Next, create an empty vector called
Bonus challenge
- Write a for loop that goes through the
nutrientsdata by row and prints out the result of a conditional statement testing whether or not dissolved inorganic nitrogen (the sum of nitrate, ammonium, and nitrite) is greater than 12. Use thedplyrfunctionslice, which lets you select rows of a data frame by their order. (Tryslice(nutrients, 2)with several numeric values, and look at the help file forslicefor more information on this function.)